What Are the FAIR Principles - and why do they matter?
Imagine having to reinvent the wheel for every scientific question—this is still the reality in much of health research today. In today's health research, data is the foundation of progress. Yet, many valuable datasets remain difficult to locate, poorly documented, or incompatible with other sources. This is where the FAIR principles, introduced in 2016 in Scientific Data by Wilkinson et al., come into play. They outline four key guidelines to optimize research data management:
FAIR stands for:

Importantly, FAIR does not automatically mean "open." Data can be FAIR-compliant while remaining restricted, as long as it is well-structured, documented, and responsibly managed.
The Four FAIR Criteria Explained
Findable: Making Data Discoverable
Let’s start with the basics: how do you find data if you don’t even know it exists? Research data only becomes valuable when it can be found. To ensure this, datasets receive globally unique identifiers (GUIDs), making each dataset distinct and easily identifiable. Machine-readable, structured metadata helps search engines and researchers locate relevant information efficiently. For example, the EU COVID-19 Data Portal assigns Digital Object Identifiers (DOIs) to datasets, linking them directly to related studies.
Findable in short:
- Globally unique identifiers (GUIDs)
- Rich, machine-readable metadata
- Indexing in searchable repositories
Accessible: Ensuring Reliable Access
Data that cannot be accessed loses much of its immediate value. FAIR encourages the use of standardized, secure protocols like HTTPS and APIs to allow reliable and transparent data retrieval. Sensitive health data often require tiered access models, such as role-based permissions or data use agreements. Use case: The German NFDI4Health consortium demonstrates successful implementation of these access controls while complying with the GDPR.
Key requirements repeated:
- Secure APIs or protocols (e.g., HTTPS)
- Persistent access to metadata, even if datasets are removed
- Built-in authentication and authorization where needed
Interoperable: Enabling Systems to Work Together
Think of interoperability as a universal language for data. It ensures that different datasets can “talk” to one another across systems and disciplines. Data reaches its full potential when it can be seamlessly integrated across different systems. This is achieved through controlled vocabularies such as SNOMED CT and LOINC, as well as standardized formats like RDF and FHIR. The OMOP Common Data Model allows institutions worldwide to harmonize their data for multinational studies. Interoperability essentially serves as a shared language that bridges system boundaries.
Remember:
- Controlled vocabularies (e.g., SNOMED CT, RxNorm, LOINC)
- Shared data models and formats (e.g., RDF, OMOP CDM)
- Metadata standards approved by scientific communities (e.g., HL7 FHIR, ISO/IEC 11179, DICOM)
Practical example: The OMOP Common Data Model (https://www.ohdsi.org/data-standardization/the-common-data-model/) allows data harmonization across health institutions, enabling global collaborative studies [5].
Reusable: Securing Long-Term Usability
Reusability is the crown jewel of FAIR. It's about enabling data to live beyond its original purpose, to fuel new hypotheses and secondary research, meaning to enable data reuse for future research. This requires clear usage licenses, thorough documentation of data provenance, and adherence to domain-specific standards like FHIR in healthcare. The U.S. All of Us program exemplifies this approach by providing standardized data spanning genomics and behavioral sciences for diverse research purposes.
Requirements in short:
- Clear usage licenses
- Detailed provenance
- Compliance with discipline-specific standards (e.g., FHIR in healthcare)
Why FAIR Matters in Health Research
Health data isn’t just big, but also costly to collect. FAIR principles help maximize its value by reducing redundant data collection and enhancing transparency through detailed metadata. A 2023 study by NFDI4Health showed that FAIR-compliant data management processes can lower data collection costs by up to 20%. These principles also facilitate international collaboration. Funding bodies like the German Research Foundation (DFG) strongly recommend depositing research data in FAIR-compliant repositories, even though this is not yet mandatory.
FAIR vs. Open Data
FAIR and Open Data often go hand in hand, but they are not the same. Open Data is about free availability; FAIR is about usability.
Clarifications:
- FAIR data may require login or ethical approval.
- Open data doesn’t always meet FAIR quality standards.
Policy update: Horizon Europe enforces FAIR principles but leaves data openness to context-sensitive decisions [8].
Germany's Leadership in FAIR Implementation
Germany has become one of the leading nations driving FAIR adoption, with several national programs and institutions working in close alignment:
- NFDI4Health (National Research Data Infrastructure for Personal Health Data): As part of Germany's broader NFDI initiative, NFDI4Health focuses on harmonizing health data across research disciplines, public health institutions, and clinical care. The project integrates epidemiological, clinical, and public health data into standardized, FAIR-compliant infrastructures. By 2025, NFDI4Health has harmonized over 500 datasets and established robust governance frameworks ensuring GDPR compliance, ethical oversight, and interoperability.
- HiGHmed (Medical Informatics Initiative): The HiGHmed Consortium demonstrates practical clinical applications of FAIR principles by connecting university hospitals and research centers across Germany. Using standards like OMOP and FHIR, HiGHmed enables secure and interoperable data exchange between clinical care and biomedical research. It has already supported numerous collaborative studies in oncology, infectious diseases, and rare disorders while maintaining strict patient privacy safeguards.
- German Research Foundation (DFG): The DFG has established guidelines promoting FAIR data management as part of good scientific practice. While not mandatory, DFG funding proposals increasingly emphasize transparent data management plans, encouraging widespread adherence to FAIR principles across German academia.
These coordinated national efforts have positioned Germany as a pioneer in building FAIR-compliant data ecosystems that bridge clinical care, public health, and biomedical research.
Global Adoption: FAIR on the Rise Worldwide
Beyond German initiatives, several international projects are setting benchmarks for successful FAIR implementation:
- NIH STRIDES (USA): The U.S. National Institutes of Health launched the STRIDES Initiative to leverage commercial cloud providers for scalable, FAIR-compliant data storage and computing. This program has enabled thousands of researchers to access petabytes of biomedical data securely, accelerating breakthroughs in genomics, cancer research, and epidemiology.
- GA4GH (Global Alliance for Genomics and Health): The GA4GH brings together hundreds of organizations worldwide to develop interoperable standards for genomic and clinical data sharing. Its Framework for Responsible Sharing of Genomic and Health-Related Data has been widely adopted, facilitating international collaboration in areas such as rare disease research, cancer genomics, and federated data analysis.
- ELIXIR Europe: ELIXIR connects life science resources across Europe, offering training, standards, and infrastructure that embed FAIR principles into day-to-day research. By 2025, ELIXIR has successfully integrated FAIR data management into dozens of national research infrastructures and offers extensive training programs that have educated thousands of scientists on FAIR data stewardship.
- EOSC (European Open Science Cloud): While still evolving, EOSC aims to create a trusted environment for FAIR data sharing across disciplines and countries. Its 2030 roadmap envisions FAIR-by-default policies for all publicly funded European research, backed by coordinated governance and infrastructure investments.
These examples highlight not only the global commitment to FAIR principles but also demonstrate concrete progress in developing technical solutions, governance models, and human capacity to make FAIR a reality.
FAIR and Machine Learning in Healthcare - the Foundation for AI in Healthcare?
The preparation of data for AI model training is one of the most labor-intensive and critical steps in healthcare AI development. Long before a model is trained, data scientists thoroughly assess data completeness, consistency, and structure. In this context, FAIR-compliant data provide a significant advantage: by ensuring high-quality, well-documented, and interoperable datasets from the outset, FAIR dramatically reduces the time and resources needed for data cleaning and transformation.
This leads to several tangible benefits:
- Minimizing AI bias by offering comprehensive and harmonized datasets.
- Enabling reproducible model development thanks to transparent provenance and standardized formats.
- Supporting federated learning approaches where data remains securely within local institutions while contributing to global model training.
A practical example is provided by GA4GH's federated learning projects, which leverage FAIR principles to enable privacy-preserving international AI collaborations across healthcare systems.
Challenges to Implementation
Turning FAIR from vision into practice comes with its own set of hurdles—technical, cultural, and ethical. For healthcare institutions, these challenges are amplified by strict regulations, legacy IT systems, and varying data literacy across teams.
Technical Barriers
FAIR implementation often begins with infrastructure. Yet many health institutions operate with outdated databases and siloed storage systems. These setups are not FAIR-ready and require heavy lifting to integrate persistent identifiers, metadata layers, and interoperable standards.
What’s needed:
- Modern, cloud-based repositories
- APIs that support FHIR, OMOP, or RDF standards
- Data lakes that allow metadata indexing and secure linkage across sources
Organizational Resistance
Even with the right tech, human factors slow adoption. Researchers may fear being "scooped" if they share data, or simply lack the time and resources to structure datasets FAIRly.
Solutions:
- Introduce FAIR compliance as a funding prerequisite
- Provide incentives and recognition for data sharing
- Employ dedicated data stewards in research teams
Ethical Complexity
FAIR data must also be ethical data. This means protecting patient privacy while maximizing utility.
Emerging best practices:
- Dynamic consent systems that allow granular control of data sharing
- Secure computation methods (e.g., federated learning)
- Privacy-preserving data linkage models
Helpful tools: FAIR-Checker (https://fair-checker.france-bioinformatique.fr) lets institutions assess and improve their FAIR maturity [11].
Future Directions - a look ahead
FAIR is not an endpoint—it’s a foundation for a new era of global health research:
Building on it, health research can embrace truly collaborative, patient-centric, and tech-forward science. Here's where the next decade is headed:
1. FAIR-by-Default in Public Research
Policy Development: Funding agencies worldwide are moving toward mandates where publicly funded datasets must be FAIR, unless justified otherwise. Germany's NFDI, the European EOSC, Australia’s ARDC, and Japan’s NBDC are laying the groundwork for enforceable FAIR standards.
2. FAIR in Academic Curricula
Next-Generation Training: Universities across Europe, North America, Asia, and Australia are embedding FAIR into medical and data science education. Practical modules on metadata standards, data licensing, and ethics prepare students for real-world research applications.
3. Citizen Science with FAIR Foundations
Citizen Science and Patient Involvement: Patient groups and citizen scientists globally contribute data through platforms like Open Humans and national biobanks. With FAIR frameworks, this data becomes interoperable and valuable for both advocacy and scientific discovery.
4. FAIR Meets AI Transparency
AI Transparency: Explainable AI is crucial in medicine. FAIR-compliant datasets simplify the tracing of data provenance and model inputs, which is increasingly essential for regulatory approvals and clinical trust.
5. Global Federated Research Ecosystems
Global Federated Networks: From rare disease registries to climate-health research, federated data architectures powered by FAIR principles allow researchers worldwide to collaborate across borders without moving sensitive data physically.
Vision: A world where a PhD student in Heidelberg, a clinician in Accra, and a health policy advisor in Tokyo, and a genomics researcher in Sydney all query the same federated database—with respect for privacy, equity, and scientific integrity.
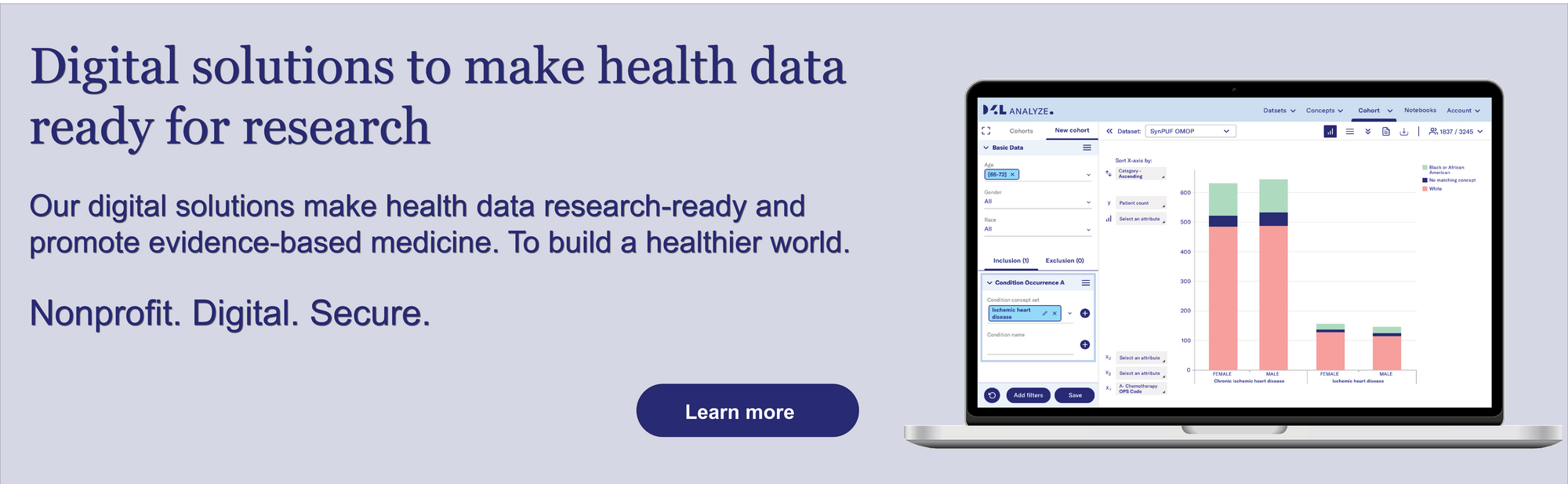
Our Solutions: FAIR at Data4Life
At Data4Life, we actively contribute to a FAIR data ecosystem with two core solutions:
D4L FAIR – A Metadata Catalog: This centralized, machine-readable catalog helps researchers identify and understand available datasets. It provides rich metadata, persistent identifiers (e.g., DOIs), and licensing information, ensuring each dataset is easily discoverable and well-documented for future use.
D2E – Federated OMOP Platform: Our "Data to Evidence" (D2E) platform enables privacy-preserving secondary research on harmonized OMOP data. D2E supports federated queries, allowing researchers across institutions to analyze data collaboratively—without the data ever leaving its secure source.
Together, these tools make FAIR a lived reality—supporting ethical, efficient, and impactful health research.
Glossary of Key Terms
- GUID (Globally Unique Identifier): Unique identifier assigned to datasets.
- RDF (Resource Description Framework): Standard for linking distributed data sources.
- FHIR (Fast Healthcare Interoperability Resources): Standard for clinical data interoperability.
- OMOP CDM (Observational Medical Outcomes Partnership Common Data Model): Model for harmonizing medical research data.
- Federated Learning: AI approach where algorithms are trained locally without sharing sensitive data.
- DOI (Digital Object Identifier): A persistent, unique identifier used to cite and link digital research objects such as datasets or publications.
The contents of this article reflect the current scientific status at the time of publication and were written to the best of our knowledge. Nevertheless, the article does not replace medical advice and diagnosis. If you have any questions, consult your general practitioner.
Originally published on